Working to tackle antibiotic resistant bacteria
Researchers at the University of Liverpool are working to tackle an increasingly antibiotic resistant bacterium.
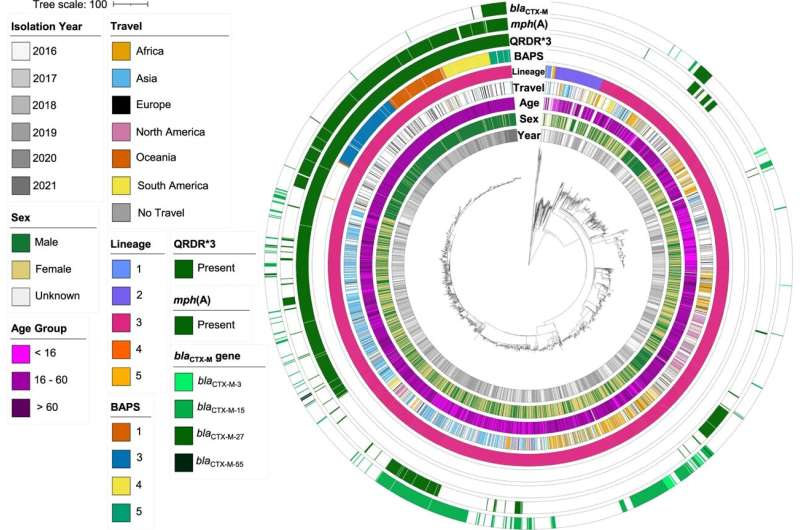
The UK Health Security Agency has reported a UK increase in this sexually transmissible bacterium, which has also been identified in countries across Europe. The bacterium, called Shigella sonnei, can cause severe diarrhea and the strain of interest is resistant to the majority of antibiotics recommended for its treatment.
Experts at the University’s National Institute for Health and Social Care Research’s Health Protection Unit in Gastrointestinal Infections led an international collaboration between scientists and public health workers from the UK, France, Belgium, Australia, and the United States of America. Together they investigated the genetic makeup and international spread of the bacterium in order to better understand the drivers behind the epidemic and predict and prevent future outbreaks.
By analyzing samples of the bacteria found in all the collaborative countries, the team developed greater insight into this antimicrobial resistant outbreak, which disproportionality affects gay, bisexual, and other men who have sex with men (MSM).
Lewis Mason, a Medical Microbiology Ph.D. Student, University of Liverpool said, “Antimicrobial resistance (AMR) is an urgent global public health crisis and our work studying this sexually transmissible intestinal infection played a key role in understanding and addressing the issue of AMR. Through this work we have learnt of the power of collaboration and the progress we’ve made shows how by cooperating and sharing genomic data we can better detect emerging disease threats around the globe.”
Kate Baker, Professor of Applied Microbial Genomics, University of Liverpool says, “Combining pathogen genomic data from across international surveillance systems allows us to pick up the spread of new strains much earlier than previously possible. We saw this with COVID as well and now we need to build on that to make the most of pathogen genomics for tackling antimicrobial resistance.”
The paper is published in the journal Nature Communications.
More information:
Lewis C. E. Mason et al, The evolution and international spread of extensively drug resistant Shigella sonnei, Nature Communications (2023). DOI: 10.1038/s41467-023-37672-w
Citation:
Working to tackle antibiotic resistant bacteria (2023, May 2)
retrieved 2 May 2023
from https://phys.org/news/2023-05-tackle-antibiotic-resistant-bacteria.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no
part may be reproduced without the written permission. The content is provided for information purposes only.
For all the latest Science News Click Here
For the latest news and updates, follow us on Google News.

