Visualizing PET’s degradation by bacterial enzymes
The rigidity, transparency and hardness of PET (Polyethylene Terephthalate) make it one of the most valuable plastics for the manufacture of plastic bottles, packaging and other single-use products. However, these characteristics make it highly persistent in the environment, to the point that a plastic PET bottle may take several hundred years to degrade in the ocean.
At the molecular level, PET, and all plastics, have a polymeric structure made up of tens of thousands of repetitions of small subunits called monomers. In the last decades, the degradation of PET by a kind of bacterial enzymes called polyester hydrolases (or PETases) has been regarded with much hope, as it is considered as a potential eco-friendly method for recycling plastic waste and recover the original monomers, thus enabling an effective circular-material economy loop.
Now, a new study led by the Institut de Ciències del Mar (ICM-CSIC) and the University of Leipzig (Germany) has revealed the details, at the molecular level, of the PET degradation process by these enzymes.
“The results of our work can be very useful for the industry, as this is the first time we can ‘see’ the motion picture of the process. Also, they pave the way to design new enzymes capable of breaking down the plastic into its original soluble components with high efficiency,” explains Francesco Colizzi, a leading author of the study.
From her side, Ania Di Pede-Mattatelli, one of the work co-authors, adds that “these enzymes could also be applied to treat PET microplastics from washing of microfleece textiles that end up in sewage treatment plants, thus contributing to the preservation of the marine environment.”
Experiments and 3D simulations
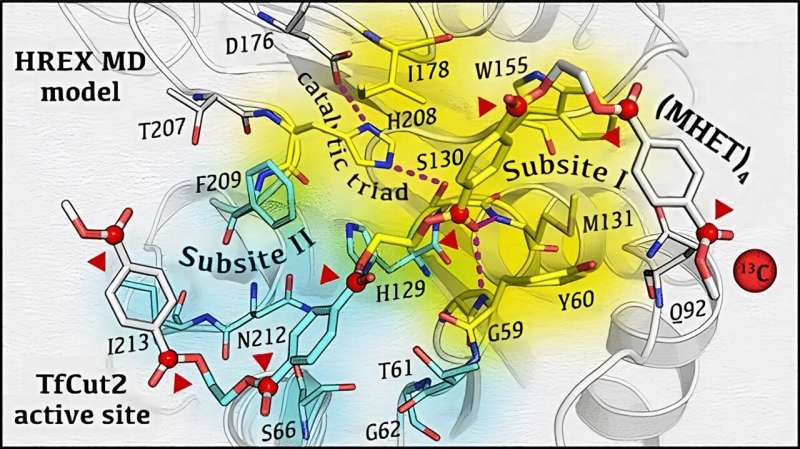
To unravel the inherent mechanism of biocatalytic degradation of PET at the atomic level, the authors of the work, recently published in the journal ACS Catalysis, designed a glass matrix that stabilized the enzymatic reaction intermediates and allowed their detection in real time by specific experiments of magnetic resonance spectroscopy. Then, using molecular calculations on a supercomputer, they were able to interpret the spectroscopic data and generate a detailed 3D molecular model of the enzymatic process of PET degradation.
Until now, how PET could bind and interact with these enzymes have been the subject of intense research, and controversial hypotheses have been put forward. For example, the simultaneous binding of a large portion of PET to the enzyme was thought to be necessary for the enzyme to break down the plastic polymer into its original components.
Instead, this work shows that the interaction of only 2 PET subunits is enough for the enzyme to cut the polymer. Lastly, the study reveals that the enzyme can “walk” or slide on the PET chain to move from one cut to the other.
“Understanding how PET interacts with the enzyme is important to guide the design of new improved systems for recycling. In the end, nature itself provides us with the starting material to reduce plastic pollution, but we must use them appropriately,” concludes Colizzi.
More information:
Patricia Falkenstein et al, On the Binding Mode and Molecular Mechanism of Enzymatic Polyethylene Terephthalate Degradation, ACS Catalysis (2023). DOI: 10.1021/acscatal.3c00259
Citation:
Visualizing PET’s degradation by bacterial enzymes (2023, May 12)
retrieved 12 May 2023
from https://phys.org/news/2023-05-visualizing-pet-degradation-bacterial-enzymes.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no
part may be reproduced without the written permission. The content is provided for information purposes only.
For all the latest Science News Click Here
For the latest news and updates, follow us on Google News.

