Study unravels the structure of bacterial P pili
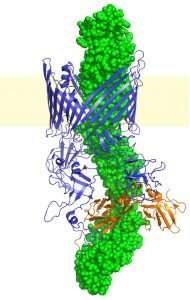
A research team led by David Thanassi, Ph.D., of Stony Brook University, has used molecular biology and cryoelectron microscopy to successfully unravel the structure of bacterial appendages called P pili. These pili are deployed by uropathogenic strains of Escherichia coli bacteria that cause kidney infections. The structure of P pili had been elusive to scientists for many years. The finding, published in Nature Communications, is a key step in order to target P pili in the infection process.
“Given their central role in initiating and sustaining infection, there is intense interest in understanding the mechanisms of pilus assembly and function,” says Thanassi, lead author and Zhang Family Professor and Chair of the Department of Microbiology and Immunology at the Renaissance School of Medicine at Stony Brook University. “Our report of the structure of P pili may help to pave the way for a method to interfere with pilus assembly or function in order to develop novel therapeutics as a potential alternative to antibiotics. A new approach may help us to better treat urinary tract infections and other infectious diseases.”
Bacterial infections of the urinary tract are common and associated with high rates of antibiotic resistance, which is a reason why Thanassi and the research team are focusing on pili structures involved in these infections. Kidney infections themselves pose a major risk for the bacteria to enter the bloodstream, an action that could result in lethal bacterial sepsis.
This latest work builds on previous research from the team and other scientists that revealed the structure of related bacterial appendages, termed type 1 pili, which facilitate colonization of the bladder.
Researchers find new lead for disarming antibiotic-resistant bacteria
Minge Du et al, Processive dynamics of the usher assembly platform during uropathogenic Escherichia coli P pilus biogenesis, Nature Communications (2021). DOI: 10.1038/s41467-021-25522-6
Citation:
Study unravels the structure of bacterial P pili (2021, September 22)
retrieved 22 September 2021
from https://phys.org/news/2021-09-unravels-bacterial-p-pili.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no
part may be reproduced without the written permission. The content is provided for information purposes only.
For all the latest Science News Click Here
For the latest news and updates, follow us on Google News.

