Study uncovers mechanisms of protein misfolding linked to neurodegenerative diseases
A team at UT Southwestern has developed a computational approach to uncover mechanisms of protein misfolding linked to neurodegenerative diseases. The study, published in Nature Communications, offers key insights that could help identify new treatments for patients.
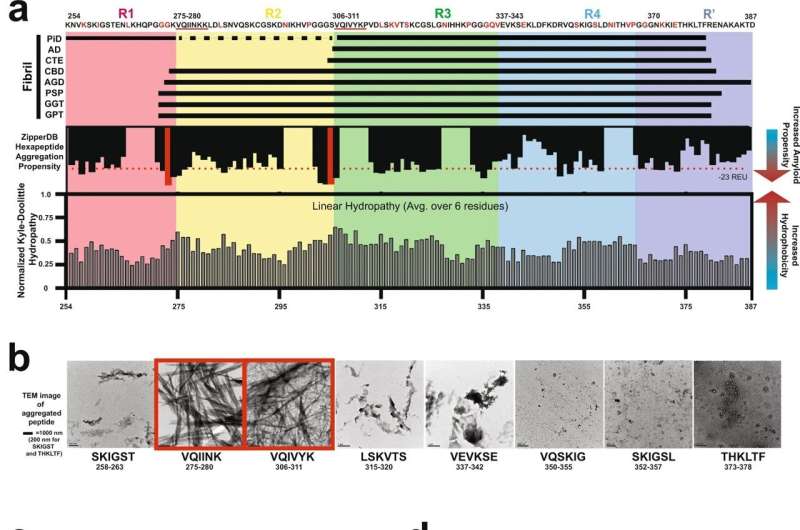
The study analyzed structures of amyloid fibrils, which are made up of proteins that are normally soluble but have assembled in a way that makes them insoluble and often dangerous. Many types of amyloids are associated with different neurodegenerative diseases.
“Understanding the mechanisms of amyloid folding will help identify novel methods to treat protein misfolding diseases,” said Lukasz Joachimiak, Ph.D., Assistant Professor of Biochemistry and in the Center for Alzheimer’s and Neurodegenerative Diseases at UTSW’s Peter O’Donnell Jr. Brain Institute. Dr. Joachimiak is an Effie Marie Cain Scholar in Medical Research and the lead author of the study.
According to Dr. Joachimiak, tau proteins play key roles in healthy brain cells. However, when a tau protein misfolds in a way that exposes certain motifs that allow it to self-replicate and build on itself, it wreaks havoc in the brain. This assembly of proteins creates distinct shapes of aggregated tau proteins that are associated with neurodegenerative diseases known as “tauopathies.” The study looked at conformations from diseases classified as tauopathies, including Alzheimer’s and chronic traumatic encephalopathy, or CTE.
Using tau as a model protein, the computational approach developed by the researchers allowed them to estimate the energetics of interactions in structures of amyloid fibrils. By applying this to 27 distinct tau protein structures, they uncovered networks of interactions involved in stabilizing different amyloid fibril folds.
This information was used to classify different tauopathies based on fibril structure conformation, opening the door to design tau sequences that only adopt a single conformation. The study is believed to be the first to computationally model the effect that mutations have on fibril structure stability—an insight that will be essential in developing methods to predict these structures from the protein sequence alone.
“We are applying these methods to control folding of other amyloid-forming proteins that direct normal biological processes to begin to manipulate them,” Dr. Joachimiak said.
The study links the team’s previous work that focused on the process of misfolding in an individual tau molecule with end-stage amyloid fibril conformations observed after onset of the disease.
More information:
Vishruth Mullapudi et al, Network of hotspot interactions cluster tau amyloid folds, Nature Communications (2023). DOI: 10.1038/s41467-023-36572-3
Citation:
Study uncovers mechanisms of protein misfolding linked to neurodegenerative diseases (2023, March 27)
retrieved 27 March 2023
from https://phys.org/news/2023-03-uncovers-mechanisms-protein-misfolding-linked.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no
part may be reproduced without the written permission. The content is provided for information purposes only.
For all the latest Science News Click Here
For the latest news and updates, follow us on Google News.

