Researchers uncover physics involved in a key process in Huntington’s disease
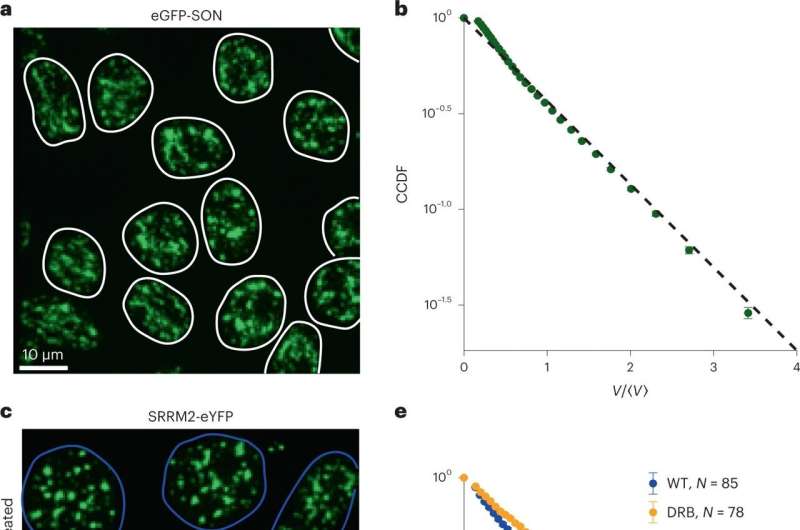
Researchers from Princeton University have uncovered the physics of a cellular process linked to aggregation diseases, including Huntington’s disease, paving the way to a deeper understanding of neurodegenerative disorders at the molecular level.
Many critical systems within a cell function inside liquid droplets that are separated from their surrounding fluids the way oil gathers in water. Scientists have only recently realized the importance of these droplets, especially in the kinds of processes that lead to cell breakdown and death.
The Princeton-led team found that as such droplets drift in their surrounding liquid, they collide and gather into larger droplets in mathematically predictable ways. The math that describes this aggregation is similar to how internet memes go viral or scientific papers accumulate large numbers of citations. Abnormally large droplets in brain cells are linked closely to ALS, Alzheimer’s and a range of other dysfunctions.
The work was led by Princeton faculty members Ned Wingreen, the Howard A. Prior Professor in the Life Sciences, and Cliff Brangwynne, the June K. Wu ’92 Professor in Engineering. Brangwynne described the work as “important for establishing a quantitative framework for understanding the fundamental physical mechanism behind protein aggregation, a necessary step for developing treatments for these devastating diseases.”
The paper is published in the journal Nature Physics.
More information:
Daniel S. W. Lee et al, Size distributions of intracellular condensates reflect competition between coalescence and nucleation, Nature Physics (2023). DOI: 10.1038/s41567-022-01917-0
Citation:
Researchers uncover physics involved in a key process in Huntington’s disease (2023, February 2)
retrieved 2 February 2023
from https://phys.org/news/2023-02-uncover-physics-involved-key-huntington.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no
part may be reproduced without the written permission. The content is provided for information purposes only.
For all the latest Science News Click Here
For the latest news and updates, follow us on Google News.

