Researchers describe a twisted cell–cell adhesion molecule complex structure
Our bodies comprise different tissues and organs, which are composed of many cells that must adhere to form functional higher order structures. This adherence is facilitated by specialized proteins called cell–cell adhesion molecules, which extend from neighboring cells and link to each other.
One class of cell–cell adhesion molecules is the Cadherin EGF LAG seven-pass G-type receptor (CELSR) cadherins. These proteins play important roles in forming tissues that have a specific shape and pattern of cells. To date, we do not fully understand CELSR cadherin structure and function.
Shigetaka Nishiguchi of ExCELLS, Kasai S. Rinshi of iGCORE at Gifu University, and Takayuki Uchihashi of ExCELLS and Nagoya University utilized single-molecule fluorescence microscopy and high-speed atomic force microscopy (HS-AFM) to elucidate the mechanism of CELSR cadherin dimerization.
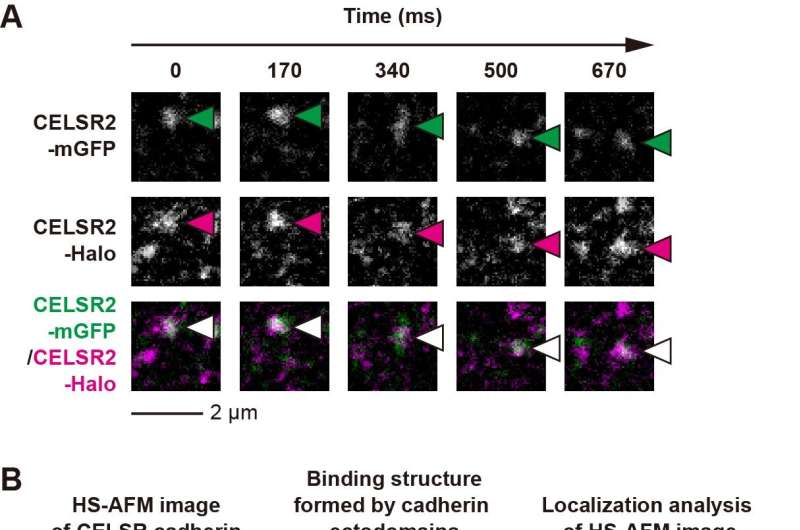
Researchers constructed two distinct fluorescently tagged CELSR cadherins (CELSR2-mGFP and CELSR2-Halo) and observed the interfaces between neighboring cells expressing CELSR2-mGFP or CELSR2-Halo using single-molecule fluorescence microscopy (Fig. 1A).
Their results demonstrated that colocalization fluorescence spots of two differently tagged CELSR cadherins at cell–cell interfaces were sustained above the long distances (~1 μm) and time (~1 sec), indicating that CELSR cadherins form specific and stable dimers to connect neighboring cells to form a minimal adhesion unit.
Next, they used HS-AFM to obtain high-resolution images of purified CELSR cadherins in solution at the nanometer scale (Fig. 1B). The images revealed that the extracellular domains (ectodomains) of CELSR cadherin that connect cells exhibited strand- and globule-like portions composed of cadherin ectodomains and non-cadherin ectodomains, respectively.
HS-AFM also revealed that the two CELSR cadherins bind through strand-like structures in an antiparallel orientation, consistent with the binding complex of CELSR cadherins between cells observed using single-molecule fluorescence microscopy. These researchers applied localization analysis of HS-AFM images previously developed by other researchers to improve the spatial resolution of their HS-AFM images and obtain higher-resolution images of the structure.
Their analysis showed that the strand-like portions of the eight cadherin ectodomains of the CELSR cadherins overlapped entirely in a twisted manner. These researchers have also demonstrated that the fourth cadherin ectodomain from the membrane-distal side of CELSR cadherin is especially important for CELSR cadherin interactions using a bead aggregation assay, which can evaluate CELSR cadherin binding activity.
Interestingly, the overlapping of eight cadherin ectodomains in the binding complex reported in this study is the most extensive interaction described to date relative to other cadherin members, and the CELSR cadherin complex is larger (~66.7 nm) than the typical extracellular space distance in the cell–cell adhesion area of epithelial cells (15–25 nm).
Researchers have hypothesized that the large binding complex formed by CELSR cadherins may act as a physical spacer between cells, allowing space for small extracellular vesicles containing messenger molecules, and regulating polarity-dependent tissue formation, which directly impacts the patterning of cells.
These findings provide new insights into the complex mechanisms by which CELSR cadherins mediate cellular adhesion. Although further detailed analyses are necessary to fully understand the physiological role of the binding structure of the CELSR cadherins reported in this study, the results have important implications for the development of new therapies for diseases involving impaired cell–cell adhesion.
More information:
Shigetaka Nishiguchi et al, Antiparallel dimer structure of CELSR cadherin in solution revealed by high-speed-atomic force microscopy, Proceedings of the National Academy of Sciences (2023). DOI: 10.1073/pnas.2302047120
Citation:
Researchers describe a twisted cell–cell adhesion molecule complex structure (2023, April 27)
retrieved 27 April 2023
from https://phys.org/news/2023-04-cellcell-adhesion-molecule-complex.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no
part may be reproduced without the written permission. The content is provided for information purposes only.
For all the latest Science News Click Here
For the latest news and updates, follow us on Google News.

