Researchers build cell atlas using scattered single-cell datasets
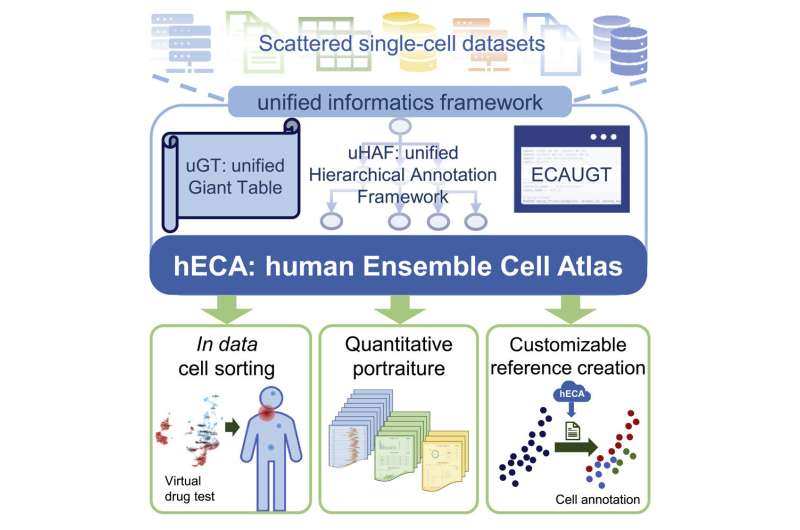
Imagine a virtual human body, rich in complexity and detail, that enables scientists to simulate experiments that can’t be conducted in vivo or in vitro. A team of Chinese researchers brought this vision closer to reality by developing a framework for seamless cell-centric data assembly and built the human Ensemble Cell Atlas (hECA) using data collected from scattered public datasets.
They presented their unified informatics framework in a study published April 28 in iScience. hECA has also made a landmark contribution to integrating human single-cell data from multiple sources and performing downstream analysis, which published in Quantitative Biology on July 4.
“Case studies of the hECA demonstrated the revolution that such a cell-centric ensemble cell atlas can bring to biomedical research,” said study author Xuegong Zhang from Tsinghua University.
The rapid development of single-cell sequencing technologies, especially an RNA-sequencing method known as single-cell transcriptomics, has allowed scientists to profile individual cells and examine which genes are switched on in different types of cells.
Scientists around the world are engaged in building single-cell-resolution “atlases” of all the different cell types in projects such as the Human Cell Atlas (HCA) and the Human BioMolecular Atlas Program. But there is still some uncertainty about how a cell atlas should be defined and assembled.
“The key point of cell atlas assembly is the organization of cell information,” Zhang said.
Since the launch of the HCA project in 2017, many papers about cell atlases have been published, and most of them are collections of a large variety of single-cell data documented and indexed on a project-by-project basis. Previous studies argued that cell mapping is about creating a three-dimensional skeleton of the human body and simply assembling the observed cells into their corresponding positions. However, a human body is too complex for this type of assembly.
Instead, “the assembly of a cell atlas should convey the multifaceted nature of the data and allow users to search with customized conditions among different indexing methods,” Zhang said.
In the meantime, massive amounts of single-cell transcriptomic data are pouring into the public domain from multi-institutional collaborations, generating petabytes of data covering all major adult human organs as well as key developmental or pathological stages.
To Zhang’s team, these scattered public single-cell data suggested an alternative approach to building a cell atlas: start from the bottom-up by assembling data from multiple sources.
To assemble data of this scale from multiple sources into an ensemble atlas, the researchers developed a unified informatics framework, which included a special database infrastructure for storing single-cell data with ultra-high dimensionality and volume, as well as a unified hierarchical annotation framework to make cell type labels from different datasets comparable and consistent. The researchers also designed an application programming interface to efficiently retrieve cells in the atlas.
With these technologies, the team developed three new schemes for applying the assembled atlas. First, they enabled in data cell sorting for selecting cells from the virtual human body of assembled cells using flexible combinations of logic expressions. They created a “quantitative portraiture” system for representing the complete information of genes, cell types, and organs. They also built a customizable reference creation for users to customize their references for cell type annotation tasks.
The researchers conducted a series of experiments to verify and illustrate the quality and usability of the assembled data in multiple application scenarios. Case examples included the investigation of drug off-targets—unintended biological consequences of a drug—throughout the whole body, which demonstrated the power of the ensemble cell atlas to open new possibilities in biomedical research.
According to the study, this type of in data cell sorting can reveal important organ-specific patterns and help scientists determine organs that are more susceptible to side effects of targeted drug therapy.
The researchers have developed strategies and technologies to integrate more high-quality data from other comprehensive datasets and will continue to improve and update future versions of the hECA.
More information:
Chenwei Li et al, Integrating human single-cell data from multiple sources, Quantitative Biology (2022). DOI: 10.15302/J-QB-022-0304. journal.hep.com.cn/qb/EN/10.15302/J-QB-022-0304
Sijie Chen et al, hECA: The cell-centric assembly of a cell atlas, iScience (2022). DOI: 10.1016/j.isci.2022.104318
Provided by
Higher Education Press
Citation:
Researchers build cell atlas using scattered single-cell datasets (2022, December 27)
retrieved 27 December 2022
from https://phys.org/news/2022-12-cell-atlas-single-cell-datasets.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no
part may be reproduced without the written permission. The content is provided for information purposes only.
For all the latest Science News Click Here
For the latest news and updates, follow us on Google News.

