New computational platform to study biological processes
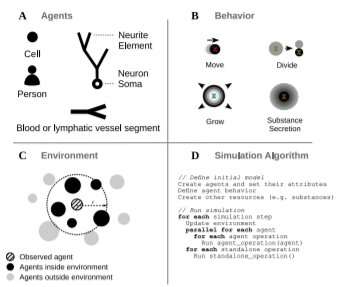
Agent-based simulations (ABS) are powerful computational tools that help scientists understand complex biological systems. These simulations are an inexpensive and efficient way to quickly test hypotheses about the physiology of cellular tissues, organs, or entire organisms. However, many ABS do not take full advantage of available computational power, and the majority of ABS platforms on the market are designed with a particular use case in mind.
In a paper published by Bioinformatics, a consortium that includes the University of Surrey, CERN, Newcastle University, GSI Helmholtz Centre, University of Cyprus, University of Geneva, SCImPULSE Foundation and Immunobrain Checkpoint, unveil their open-source, multi-disciplinary simulation platform called BioDynaMo.
In the paper, the consortium details how BioDynaMo can simulate complex medical cases in neuroscience, oncology, and epidemiology, thanks to its ability to utilize multi-core CPUs and offload computations to hardware accelerators.
The team’s results show that BioDynaMo performs up to 945 times faster than current state-of-the-art baselines. The advances make it feasible to simulate each use case with one billion cells on a single server, showcasing the potential BioDynaMo has for computational biology research.
Dr. Roman Bauer from the University of Surrey, co-founder of the BioDynaMo initiative and senior author of the study, said:
“I believe that if we are ever to tackle highly complex health issues such as Alzheimer’s, computer simulation tools like BioDynaMo will be critical for scientists moving forward.
“In this paper, we have shown that BioDynaMo’s excellent features and performance. It will help the scientists of tomorrow pitting hypotheses against each other to study their subtle or less so divergences. And more importantly, it will help biomedical research become a truly interdisciplinary field of the 21st century.”
Lukas Breitwieser et al, BioDynaMo: a modular platform for high-performance agent-based simulation, Bioinformatics (2021). DOI: 10.1093/bioinformatics/btab649
Citation:
New computational platform to study biological processes (2021, September 27)
retrieved 27 September 2021
from https://phys.org/news/2021-09-platform-biological.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no
part may be reproduced without the written permission. The content is provided for information purposes only.
For all the latest Science News Click Here
For the latest news and updates, follow us on Google News.

