New approach to transcriptomics reveals properties of 35 neuron subtypes in mice
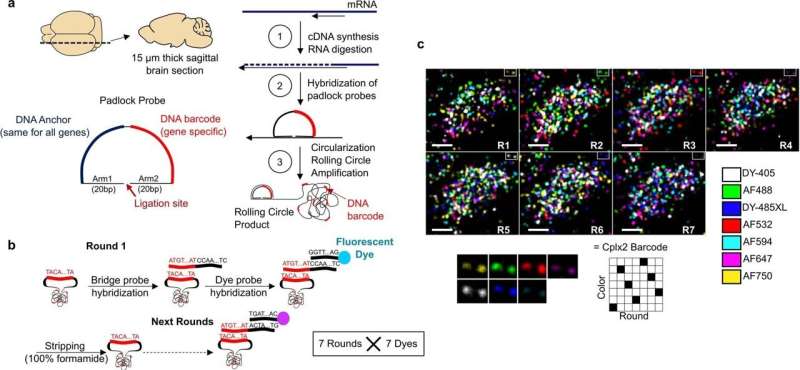
A team of researchers from University College London, Columbia University and Oxford University has developed a new approach to conducting transcriptomics and has used it to reveal the properties of 35 neuron subtypes in mice. The group has published their research in the journal Nature. Hongkui Zeng and Saskia E. J. de Vries with the Allen Institute for Brain Sciences have published a News & Views piece in the same journal issue outlining the work done by the team.
One of the goals of biological scientists is to learn how the brain works, in other animals and in humans. Considering the complexity of the brain, the goal is ambitious to be sure. But scientists believe that it can be done by carrying out a variety of step-by-step studies that seek to explain the various parts of a given brain to describe what they do. Part of that effort involves creating a catalog of brain cell types and subtypes.
One aspect of cell type classification involves deciphering and recording the repertoire of expressed genes—a process called transcriptomics. From this comes the identification of the roles that each of the individual genes play in brain circuitry. To that end, researchers have used various techniques to conduct transcriptomics (which have to be carried out on live tissue) to learn more about regions of the brain, such as the mouse visual cortex. To date 95 groups have been identified.
In this new effort, the researchers have developed a new way to carry out such work—they call it coppaFISH, and it is capable of determining the expression of 72 genes at once from a thin slice of brain tissue. It involves combining in vivo two-photon calcium imaging with conventional transcriptomic methods. The resulting expression profiles for the genes can then be used to map the transcriptomes to cell identities.
The researchers used the technique to obtain new data for 1,090 interneurons, involving 35 neuron subtypes. In so doing they found that certain transcriptomic principles could be used to predict the activity of other cell types, resulting in a transcriptomic axis that correlates with some types of cell properties. The researchers suggest their technique could be used in other regions of the brain as well.
Stéphane Bugeon et al, A transcriptomic axis predicts state modulation of cortical interneurons, Nature (2022). DOI: 10.1038/s41586-022-04915-7
Hongkui Zeng et al, A gene-expression axis defines neuron behaviour, Nature (2022). DOI: 10.1038/d41586-022-01640-z
© 2022 Science X Network
Citation:
New approach to transcriptomics reveals properties of 35 neuron subtypes in mice (2022, July 8)
retrieved 8 July 2022
from https://phys.org/news/2022-07-approach-transcriptomics-reveals-properties-neuron.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no
part may be reproduced without the written permission. The content is provided for information purposes only.
For all the latest Science News Click Here
For the latest news and updates, follow us on Google News.

