New 3D atomistic imagery of SARS-CoV-2 shows how virus uses spike protein to fuse with and infect human cells
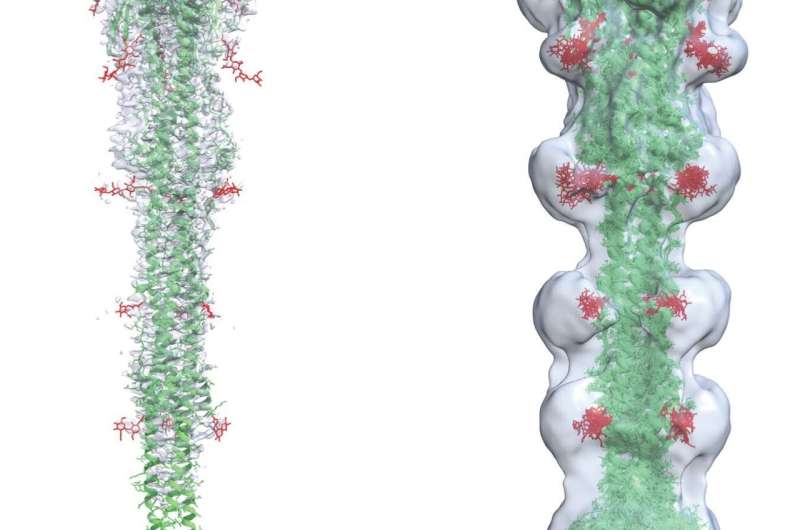
New computer models and simulations from Los Alamos National Laboratory are showing researchers how the virus that causes COVID-19 manages to use its spike protein to fuse with and infect human cells. To be presented at the March meeting of the American Physical Society, the atomistic-level imagery is highly consistent with cryo-electron microscopy data, despite the severe challenges of imaging at such high resolution.
“Better understanding of COVID-19 virus entry and structure of the spike protein will help clarify the mechanism of COVID-19 infection, the effect of variants, help with vaccine optimization, and help with drug design for treatment,” said Karissa Sanbonmatsu, a Los Alamos structural biologist on the project. Developing a digital twin of the extremely tiny but important structures allows researchers to explore potential techniques for blocking the infection at the source.
Why choose this tiny piece of the spike to study? COVID-19 infections require the virus to enter human host cells, and the spike protein plays a key role in this process, said Sanbonmatsu. In fact, the many variants of the coronavirus, including Delta and Omicron, have multiple mutations in the spike protein.
One step of virus entry is called virus-cell fusion, where the coronavirus actually fuses with the human host cell. This is not well understood, especially the region of the virus’s spike protein called the fusion peptide region. The fusion region is thought to be highly dynamic and is difficult to image at high resolution with conventional techniques such as cryo-electron microscopy and X-ray crystallography.
“Because the fusion region of the spike protein mediates virus-cell fusion and aids in virus entry into the cell,” said Chang-Shung Tung, a partner on the research, “this study provides basic mechanistic data that may be useful for understanding variants and improving vaccines and treatments.”
Using the Chicoma computing platform, one of the supercomputers at Los Alamos, Los Alamos researcher Tung created 3D structural models of this region, and Sanbonmatsu performed molecular simulations, creating an ensemble of 3D structures highly consistent with cryo-electron microscopy data, providing some of the first 3D images of the fusion region in atomistic detail.
“The spike protein undergoes many twists and turns during viral entry, making it difficult to visualize,” said Tung. “Building on data from other viruses, we used 3D modeling to capture regions of the spike people haven’t seen before.”
The paper is forthcoming, after a presentation at the March American Physical Society meeting.
Citation:
New 3D atomistic imagery of SARS-CoV-2 shows how virus uses spike protein to fuse with and infect human cells (2022, March 7)
retrieved 7 March 2022
from https://phys.org/news/2022-03-3d-atomistic-imagery-sars-cov-virus.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no
part may be reproduced without the written permission. The content is provided for information purposes only.
For all the latest Science News Click Here
For the latest news and updates, follow us on Google News.

