Genome sequencing of Moraxella bovis opens new doors for interventions against ‘pinkeye’ in cattle
Scientists have revealed that there are two different variants, or genotypes of Moraxella bovis (M. bovis), a bacterium known to cause pinkeye in cattle. This finding helps scientists understand how different types of M. bovis cause infection in livestock and can help develop preventative measures to protect U.S. cattle against this disease.
Cattle pinkeye, or bovine infectious keratoconjunctivitis, is a very contagious eye infection that causes redness, itchiness, pain, and discomfort in the eyes of affected animals. Severe cases can result in blindness and impair weight gain in calves, and thus are a concern for animal well-being and have negative economic impacts on the beef industry.
USDA Agricultural Research Service (ARS) and University of Nebraska-Lincoln (UNL) scientists sequenced and compared the genomes of a collection of M. bovis variants and found that they consisted of two major genotypes. They identified DNA differences between the genotypes. In addition, they found substances that can potentially be used as targets to control the disease.
“We found major differences in virulence factors between the two genotypes,” said Emily Wynn, ARS research microbiologist. “For example, M. bovis has a toxin, called hemolysin toxin, which it uses to penetrate the eye. We found that the two genotypes have different versions of the toxin. This difference and others among the collection of M. bovis variants could mean that there are variations in their ability to cause disease.”
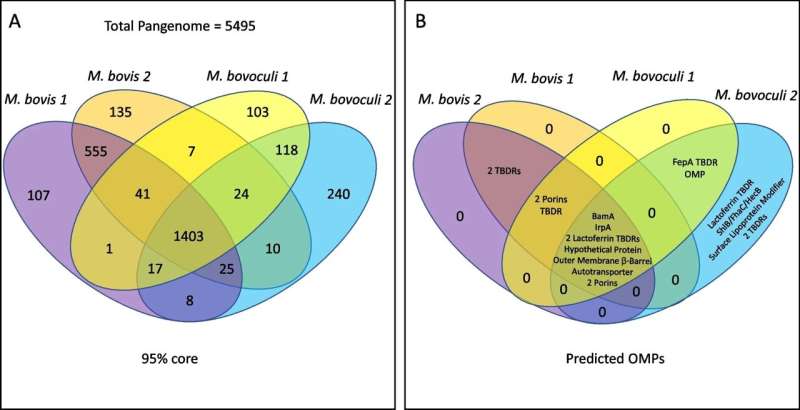
In addition, the scientists identified proteins (substances) located on the outer membrane of the bacteria cell.
The specific location of these proteins makes them available to the host immune system because they are located on the outer membrane. Proteins that are unique to one or both genotypes can be used as a target to develop specific preventative actions against any of the genotypes, if necessary,” added Wynn.
This is important because for years scientists have been closely looking at another substance in this bacterium to develop interventions against the disease, called pilin proteins. Pilins facilitate the attachment of M. bovis to the eye. However, using pilins to develop interventions could be tricky.
“The pilin gene of M. bovis can undergo an inversion,” said Mike Clawson, ARS research molecular biologist at the U.S. Meat Animal Research Center in Clay Center, NE.
“This is where parts of the gene flip and are rearranged on the bacterial chromosome. As a result, a newly formed pilin gene is created that encodes a new protein version of itself, which helps M. bovis avoid recognition by the immune system. Pilin gene inversions have been thought to be a relatively rare process. However, we saw evidence that multiple M. bovis strains were undergoing the inversions during our study. This makes the pilin gene a challenge to use as a target and emphasizes why the outer membrane proteins identified in this study are an important discovery.”
The team sequenced M. bovis strains isolated from cattle from seventeen U.S. states and one Canadian province that were assembled by Dustin Loy, UNL professor and veterinary diagnostic microbiologist.
“The first finished genome of M. bovis was produced by this collaboration on an experimental strain in 2018,” said Loy. “Since then, we haven’t seen much progress in comprehensive sequencing of this bacterium until this study between ARS and UNL.”
Loy has dedicated years of research to understanding this infectious disease, collecting samples directly from veterinarians working with cattle—for testing and identifying variabilities between the strains.
“This disease is often overlooked. Still, it is the most frequently reported disease in beef breeding cows and second in calves. Our work recognizes the economic impact this causes to beef producers,” added Loy.
The team commemorates collaborative research on pinkeye that goes back approximately 58 years with the groundbreaking work of ARS researcher George Washington Pugh Jr., who was the first black scientist in the agency, and made major advances in understanding the role of M. bovis in pinkeye. More recently, UNL published a collaborative study with ARS assessing immunological responses and the effectiveness of vaccines to protect cattle against diseases associated with these bacteria.
The study was published in BMC Microbiology.
Emily L. Wynn et al, Whole genome sequencing of Moraxella bovis strains from North America reveals two genotypes with different genetic determinants, BMC Microbiology (2022). DOI: 10.1186/s12866-022-02670-3
Citation:
Genome sequencing of Moraxella bovis opens new doors for interventions against ‘pinkeye’ in cattle (2022, October 25)
retrieved 26 October 2022
from https://phys.org/news/2022-10-genome-sequencing-moraxella-bovis-doors.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no
part may be reproduced without the written permission. The content is provided for information purposes only.
For all the latest Science News Click Here
For the latest news and updates, follow us on Google News.

