Euchromatin is not really open in living cells, shows study
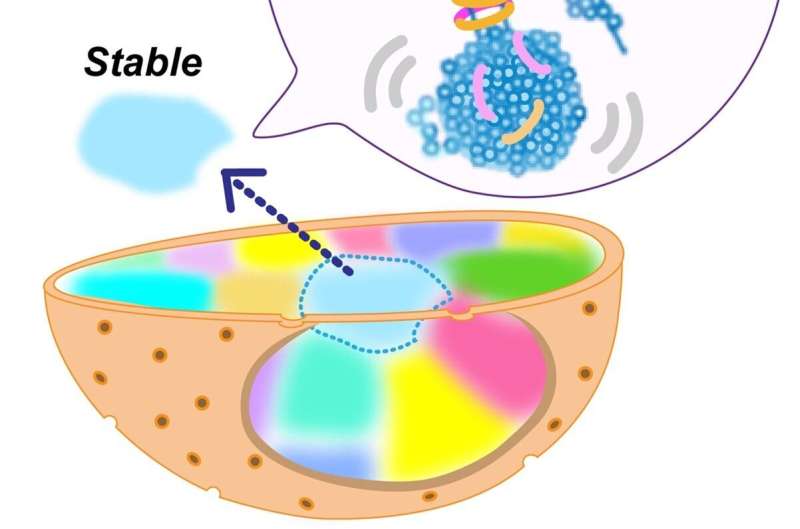
DNA and associated proteins in active regions of the genome are condensed but behave like a viscous liquid at the molecular level. This finding greatly increases our understanding of the physical nature of expressed genome regions in living human cells.
The human genome DNA has a remarkable capacity for compaction. When 46 sets of human chromosomes are stretched end to end, they collectively reach two meters in length but are somehow arranged in a nucleus with only about ten micrometers in diameter. To fit inside the nucleus, the strands of DNA are wrapped around groups of histone proteins, like a thread around a spool, to form structures known as nucleosomes. Nucleosomes can be folded with other proteins into more compact structures, called chromatin. Despite great advances in technology over the last century, the physical characteristics of actively expressed chromatin, or euchromatin, remain a mystery.
Recent research unveils that euchromatin is condensed, rather than decondensed and open, in living human cells and behaves like a liquid at the nucleosome level. At the chromosome level (or the micrometer scale), chromatin is stable and behaves like a solid, which may limit DNA damage by reducing entanglements of long DNA.
A team led by Professor Kazuhiro Maeshima of SOKENDAI and the National Institute of Genetics (Research Organization of Information and Systems) recently investigated euchromatin, to determine the physical characteristics of euchromatin, whether it is open or condensed and liquid- or solid-like. Euchromatin is often transcribed for gene expression in living human cells. The team used a combination of genomics, single-nucleosome imaging, and computational modeling to assess euchromatin in cells.
Prior to this study, euchromatin was thought to have an open, or less condensed, conformation to allow the large proteins associated with gene transcription access to genomic DNA. Instead, the researchers discovered that euchromatin in living cells formed condensed domains that behaved like a viscous fluid at the level of individual nucleosomes and behaved like a solid at the chromosome level.
The team published their results on April 5 in Science Advances.
“We demonstrated that euchromatin in living human cells does not necessarily exist as open structures, but instead it essentially forms condensed domains,” said Maeshima.
The research team introduced fluorescently-labeled nucleotides into living cells and visualized nucleosomes in close vicinity to one another using super-resolution microscopes. “Nucleosomes, the basic units of chromatin, fluctuated and behaved like a liquid inside the condensed euchromatin domains,” said Maeshima. What the scientists observed challenged conventional thinking regarding the conformation of euchromatin.
“The dynamic, liquid-like behavior of nucleosomes we observed allows active transcription even in condensed euchromatin,” said Maeshima. “The condensed domains can protect DNA from radiation damage due to reduced physical access to DNA and decreased reactive chemical production,” one of the co-authors, Shiori Iida added.
In contrast to the nucleosome level, euchromatin behaved like a solid at the chromosomal level, which the researchers propose could avoid excess entanglements of long genome DNA and keep it intact without breaks.
The new model of condensed euchromatin structure proposed by the researchers provides a mechanism to decrease the “stickiness” of chromatin through the acetylation of, or addition of an acetyl group to, histones. This opens up chromatin structure in order to accommodate large transcription or replication complexes and to regulate the expression of genes. The researchers also point out that chromatin condensation and organization may play an important role in cell differentiation, or specialization, as the chromatin domains of undifferentiated embryonic mouse stem cells, for example, are more fluid and less defined.
“Our ultimate goal is to reveal how genome information is searched and read out in living cells,” said Maeshima. The current study will help scientists better understand gene regulation, DNA replication and repair in living cells. The novel imaging techniques the research team developed will also provide a means for researchers to investigate other nanometer-scale molecules and their dynamics within the cell.
More information:
Tadasu Nozaki et al, Condensed but liquid-like domain organization of active chromatin regions in living human cells, Science Advances (2023). DOI: 10.1126/sciadv.adf1488. www.science.org/doi/10.1126/sciadv.adf1488
Provided by
Research Organization of Information and Systems
Citation:
Euchromatin is not really open in living cells, shows study (2023, April 5)
retrieved 5 April 2023
from https://phys.org/news/2023-04-euchromatin-cells.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no
part may be reproduced without the written permission. The content is provided for information purposes only.
For all the latest Science News Click Here
For the latest news and updates, follow us on Google News.

