Establishing the basis for mammalian intracellular computing with mRNA switches
Professor Hirohide Saito and his research team have developed a new approach to control mRNA translation using Cas proteins, thus expanding the variety of mRNA switches and making it easier to build synthetic gene circuits in mammalian cells. The results of this research were published in Nature Communications on April 19, 2023.
Cellular computing technologies that enable precise, computer-like control of cellular function and fate, are being explored in the field of synthetic biology. By programming gene expression patterns as desired, researchers can vastly deepen our understanding of various biological processes and create new medical treatments. In the future, this technology could help improve therapeutic efficacy and reduce side effects in drug discovery, vaccine development, cell transplantation, and other forms of medical treatment.
In order to create such precise control systems, it is necessary to construct complex circuits that process information like a computer in living cells, which in turn necessitates the use of artificial genetic circuits based on the building blocks of living systems such as DNA, RNA, and proteins. However, the availability of components amendable to control by scientists has been a limiting factor for precise programming of cellular behavior.
Recently, researchers have begun tapping into the power of RNA-binding proteins (RBPs) as translational regulators. However, this approach has been hindered by a limited number of RBPs capable of regulating translation, especially those with multiplex compatibility, to fully take off for building post-transcriptional synthetic biological circuits for basic research and clinical applications.
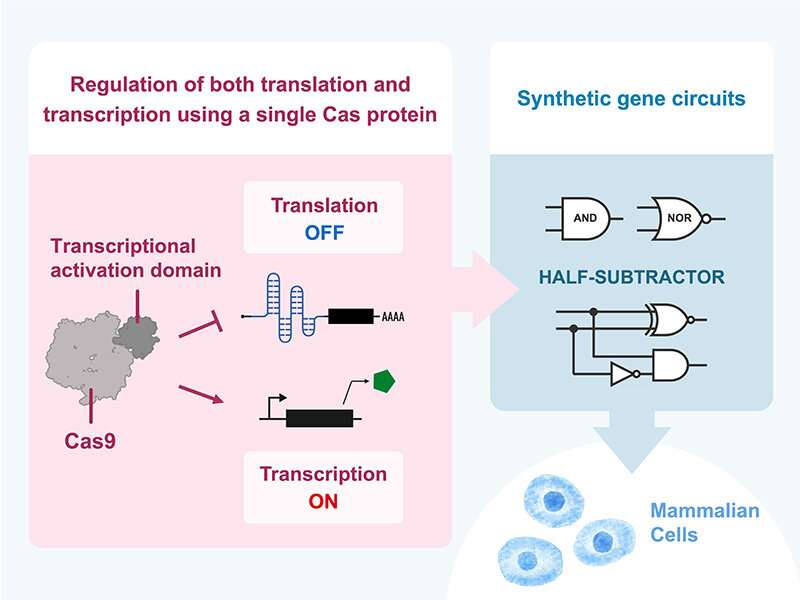
To lengthen the list of RBPs available as translational regulators, Saito and his group exploited the ability of CRISPR-Cas proteins to recognize specific sgRNA (single guide RNA) sequences by inserting them into the 5′-UTR (untranslated region) of a target mRNA that encodes a reporter gene product (switch).
In the absence of the corresponding Cas protein (trigger), the reporter gene product undergoes translation unopposed, meaning that the switch is in its ON state. If cells express the Cas protein, however, it binds to the target mRNA via the sgRNA sequence and blocks translation of the reporter gene product, thereby turning the switch to its OFF state (OFF switch: ON state → OFF state). By incorporating a so-called “switch-inverter” module into the system, the team can convert these OFF switches into ON switches (OFF state → ON state).
The research team named this translational regulation system CARTRIDGE (Cas-Responsive Translational Regulation Integratable into Diverse Gene control) since they successfully utilized bioengineering technologies for controlling Cas proteins, such as the split-Cas system and anti-CRISPR proteins, to manipulate the translation of many different mRNA switches.
In addition, the team tested 25 types of Cas proteins and found that 20 proteins can function in repressing translation and 13 proteins interacted only with their specific mRNA counterparts, which is crucial when utilizing them in combination for constructing artificial genetic circuits. Therefore, this method has increased the diversity of available mRNA switches tremendously.
With this expansive toolkit, the scientists built various complex artificial circuits, such as translational logic AND gates and multi-layered cascades. They also designed a complex half-subtractor arithmetic circuit in mammalian cells by employing both transcriptional and translational control by Cas proteins.
By vastly expanding the molecular toolkit available for translational regulation, the research team hope this work will inspire not only RNA- and RBP-based synthetic biology applications but also fuel future development of genome editing technologies and CRISPR studies, upon which CARTRIDGE was built.
More information:
Shunsuke Kawasaki et al, Programmable mammalian translational modulators by CRISPR-associated proteins, Nature Communications (2023). DOI: 10.1038/s41467-023-37540-7
Citation:
Establishing the basis for mammalian intracellular computing with mRNA switches (2023, April 24)
retrieved 24 April 2023
from https://phys.org/news/2023-04-basis-mammalian-intracellular-mrna.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no
part may be reproduced without the written permission. The content is provided for information purposes only.
For all the latest Science News Click Here
For the latest news and updates, follow us on Google News.

