Computational biologists design a novel and improved triosephosphate isomerase barrel protein
Proteins and enzymes perform several key functions inside the human body. To design a functional protein, it is important to be able to control the structure of protein folds and understand the relationship between the sequence, structure, and stability of proteins. Recent developments in computational biology have enabled the de novo design of proteins with varying folds and structures.
One such structure is the triosephosphate isomerase (TIM) barrel protein fold, which occurs in nearly 10% of all enzymes, and is involved in protein-mediated metabolism. It has a simple structure with repeating beta/alpha subunits that are connected by variable loops and is thus used widely as a scaffold to design other proteins. However, it has not been exploited completely to design functional proteins, due to challenges in altering its overall architecture.
Recently, a team of researchers led by Dr. Po-Ssu Huang from Stanford University conducted a study to investigate whether the structure of the central beta barrel could be altered de novo, while eliminating structural loops and improving its stability. Their goal was to design a TIM barrel protein with high stability and functional properties, and their findings were published in BioDesign Research.
“Although a TIM barrel protein has been designed de novo previously, it was difficult to finely alter the curvature of its central beta barrel, thus limiting its utility for functional design,” says Dr. Huang while discussing previous attempts at creating a functional protein using the TIM barrel fold.
First, the team used the RosettaRemodel (24) framework to generate and identify ideal protein backbones using an autoregressive approach. Next, they used an iterative sequence design protocol to generate multiple sequences with a high proportion of successfully folding designs.
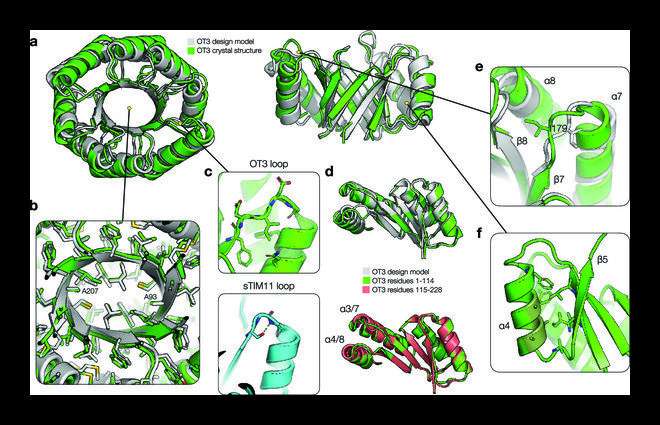
Together with protein synthesis and structure determination, a TIM barrel protein was developed de novo with two-fold (ovoid) symmetry and a completely new syntax, i.e., topological information, and a new sequence. The crystalline structure of this protein closely resembled the design model created by the team, confirming their design hypothesis.
Regarding the structural properties of the TIM barrel protein, Dr. Huang says, “The designed protein exhibited an elongated β barrel architecture with loops that were not structurally involved and a more developed hydrophobic core.”
The team further found that the designed sequences were highly stable and were able to fold to the designed barrel curvature. In addition, the shape of the ovoid TIM barrel was found to be suitable for the incorporation of different residue identities and combinations.
Further, the team employed mutagenesis—a process wherein key amino acid residues constituting a protein get replaced with amino acids that have similar or contrasting properties. Quite astonishingly, despite the modification, the resulting TIM barrel protein displayed high structural and thermal stability, although it reduced the overall yield of the protein to a certain extent.
What are the long-term implications of these findings? “Our designs show robustness to drastic mutations, retaining high melting temperatures even when multiple charged residues are buried in the hydrophobic core or when the hydrophobic core is ablated to alanine. As a scaffold with a greater capacity for hosting diverse hydrogen bonding networks and installation of binding pockets or active sites, the ovoid TIM barrel represents a major step towards the de novo design of functional TIM barrels,” says Dr. Huang.
In summary, the novel design of the TIM barrel fold has multiple implications in the field of molecular recognition and enzyme catalysis. Owing to the frequent occurrence of TIM barrel structures in key enzymes, this study also has likely therapeutic implications.
More information:
Alexander E. Chu et al, De Novo Design of a Highly Stable Ovoid TIM Barrel: Unlocking Pocket Shape towards Functional Design, BioDesign Research (2022). DOI: 10.34133/2022/9842315
Provided by
BioDesign Research
Citation:
Computational biologists design a novel and improved triosephosphate isomerase barrel protein (2022, December 14)
retrieved 15 December 2022
from https://phys.org/news/2022-12-biologists-triosephosphate-isomerase-barrel-protein.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no
part may be reproduced without the written permission. The content is provided for information purposes only.
For all the latest Science News Click Here
For the latest news and updates, follow us on Google News.

