Examining impacts of elevated salinity on microbial interactions within activated sludge microbial community
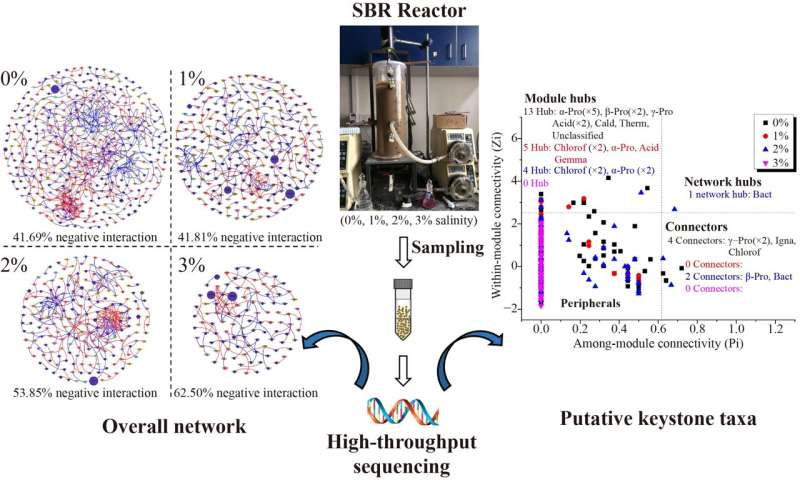
Biological treatment processes are critical for sewage purification, wherein microbial interactions are tightly associated with treatment performance. Previous studies have focused on assessing how environmental factors (such as salinity) affect the diversity and composition of the microbial community but ignore the connections among microorganisms. To fill this gap, an international team of researchers conducted an in-depth analysis of microbial interactions at elevated salinity in activated sludge systems.
Biological treatment processes are widely used in the removal of pollutants on account of their low cost and high efficiency. But high salinity could cause poor treatment performance of biological wastewater treatment plants (WWTPs), since high osmotic pressure is lethal to microbes and can inhibit the enzyme activity. Therefore, wastewater salinity is a noteworthy problem in biological treatment processes all over the world.
Activated sludge system is a complex micro-ecosystem, in which various bacterial taxa interact with each other through energy transfer and exchange of substances and information to form a large and complex ecological network to efficiently remove organic matter and nutrients. Previous studies have shown that salinity reduced the diversity of bacterial community, altered the composition of nitrifiers and denitrifiers, and further inhibited the activity of nitrifying bacteria.
The activated sludge microbial community has been investigated extensively, but seldom focuses on the interactions among microbial taxa during system operation.
It is expected that some physicochemical changes (e.g., influent salinity) might disturb the microbial interactions among various functional populations (e.g., heterotrophic bacteria, nitrifiers, denitrifiers, polyphosphate accumulating organisms (PAO), etc.), and accordingly impact the performance efficiency in activated sludge systems. However, information regarding microbial interactions and how they respond to elevated salinity has been rarely reported.
To fill these gaps, researchers from Beijing University of Chemical Technology and Beijing Technology and Business University described the microbial interactions in response to elevated salinity in an activated sludge system by performing an association network analysis. Their study reveals that higher salinity resulted in low microbial diversity, and small, complex, more competitive overall networks, leading to poor performance of the treatment process.
This study entitled “Responses of microbial interactions to elevated salinity in activated sludge microbial community” is published in Frontiers of Environmental Science & Engineering.
In this study, by examining the dynamic variation of molecular ecological networks (MENs), the research team explored the following questions: 1) How does the overall network structure respond to elevated salinity? 2) How does the subnetwork structure of different phylogenetic taxa respond to elevated salinity? 3) How do the functional bacteria and keystone species respond to elevated salinity? This study will provide a novel insight into the dynamic changes of microbial interaction under physicochemical changes.
In their study, the researchers found that 3% salinity inhibited TN removal and reduced the diversity of microbial communities. Network analysis revealed that overall networks under higher salinity conditions (2% and 3%) exhibited more complex, tighter networks, and more competitive bacterial interactions.
Subnetworks of bacteria with same function (such as AOB, NOB, and denitrifiers) differed substantially when exposed to elevated salinity. The connection between Nitrospira (NOB) and other species was seriously inhibited under 1%–3% salinity resulting in the elevation of NAR over 99.72%. In addition, the team noted that keystone species (hubs and connectors) were dynamics when exposed to different salinity and played crucial roles in maintaining system stability, despite the low abundances in microbial community.
This study successfully evaluated the effects of elevated salinity on the ecological networks of activated sludge system by performing a novel RMT-based network analysis. This work improves our understanding of the relationship between system performance and microbial interaction dynamics of activated sludge microbial communities in response to elevated salinity.
More information:
Tao Ya et al, Responses of microbial interactions to elevated salinity in activated sludge microbial community, Frontiers of Environmental Science & Engineering (2022). DOI: 10.1007/s11783-023-1660-x
Provided by
Higher Education Press
Citation:
Examining impacts of elevated salinity on microbial interactions within activated sludge microbial community (2023, June 16)
retrieved 16 June 2023
from https://phys.org/news/2023-06-impacts-elevated-salinity-microbial-interactions.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no
part may be reproduced without the written permission. The content is provided for information purposes only.
For all the latest Science News Click Here
For the latest news and updates, follow us on Google News.

