Research team develops nanosensor capable of single-molecule precision in detecting biomarkers
While it is a popular hobby for many, fishing is also a pastime full of uncertainty. Each time you have something on the line, you can never be completely sure what type of fish you’ve hooked until you pull it out of the water. In a similar way, scientists “fishing” for biomarkers—molecules whose health care applications include signaling for the presence of cancer—in biofluids such as blood can also encounter unpredictability. Finding a specific protein biomarker in a pool of thousands is like trying to catch a particular fish species in the vast ocean.
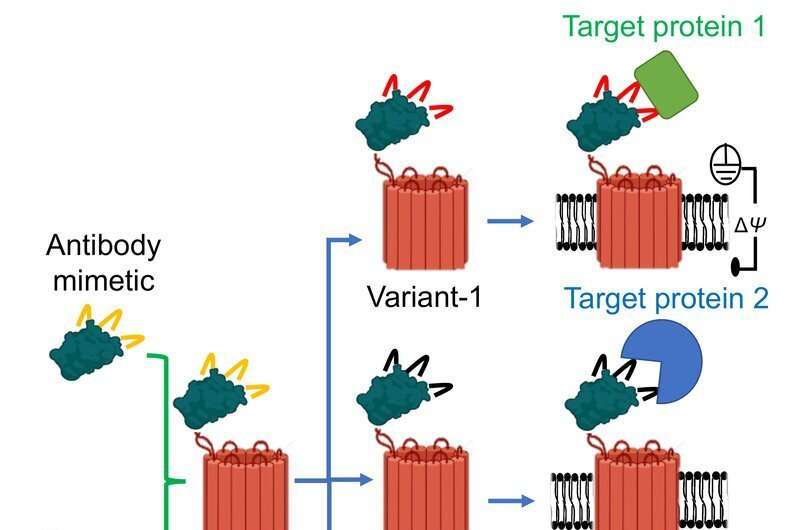
Luckily, a team of researchers from Syracuse University’s College of Arts and Sciences (A&S), SUNY Upstate Medical University, Ichor Therapeutics, and Clarkson University have devised a tiny, nano-sized sensor capable of detecting protein biomarkers in a sample at single-molecule precision. Fittingly coined as “hook and bait,” a tiny protein binder fuses to a small hole created in the membrane of a cell—known as a nanopore—which allows ionic solution to flow through it. When the sensor recognizes a targeted molecule, the ionic flow changes. This change in flow serves as the signal from the sensor that the biomarker has been found.
“These nanopores are equipped with hooks that pull certain protein biomarkers from a solution,” says Liviu Movileanu, professor of physics in A&S, who co-authored the study along with postdoctoral researcher Mohammad Ahmad. “By fishing them from the solution quickly and accurately, we can better identify and quantify protein biomarkers that are associated with various hematological malignancies and solid tumors.”
The team’s latest research, published in Nature Communications, addresses previous challenges that existed in making this technology generalizable. Their new findings formulate a sensor design architecture that can be applied to a broad range of protein targets.
Combining innovative technologies
For the first time, the team coupled nanopore technology with antibody mimetic technology—artificially designed protein scaffolds that bind and interact with a specific biomarker and behave like antibodies. Cells inside the body design their own antibodies which bind to and eliminate unwanted substances. When it comes to therapeutics, scientists engineer small proteins to penetrate cells and stimulate the production of antibodies which target specific pathogens like viruses or bacteria.
“Researchers design the scaffolds using established scaffolds from mother nature and adapt them using evolutionary mutagenesis—where they scan billions of DNA mutations until they find some that interact strongly with a specific protein,” says Movileanu. “Creating highly specific protein detection technologies will address these demands and also accelerate discoveries of new biomarkers with potential consequences for the progression of pathological conditions.”
According to Movileanu, in addition to working in a clean solution, the sensor is also highly effective in complex biofluids, like blood serum, that contain numerous antibodies.
“Essentially you have a very specific hook that targets a very specific protein,” he explains. “Since the signal encodes the exact protein that you are targeting, this technique does not have false positives, making it practical for biomedical diagnostics.”
To validate their findings, the team tested their hypothesis using a blood serum sample. With their technology, they were able to identify and quantify epidermal growth factor receptor (EGFR), a protein biomarker in various cancers. In addition, numerous calibrations of the sensors were conducted using other biophysical techniques.
At the forefront of diagnosis
While their paper provides a concept prototype, Movileanu says the project paves the way for broad applications. For example, by integrating the sensors into nanofluidic devices, this technology would allow scientists to test for many different biomarkers at once in a specimen, providing a fundamental basis for biomarker detection in complex biofluids.
“The future of medicine won’t rely as much on imaging and biopsies when diagnosing cancers,” says Movileanu. “Instead, researchers will use nanosensor technology, like what we are developing in our lab, to test blood samples for the presence of various biomarkers associated with different cancers. This research is critical to the future of prognostics, diagnostics and therapeutics.”
More information:
Mohammad Ahmad et al, A generalizable nanopore sensor for highly specific protein detection at single-molecule precision,Nature Communications (2023). DOI: 10.1038/s41467-023-36944-9. www.nature.com/articles/s41467-023-36944-9
Citation:
Research team develops nanosensor capable of single-molecule precision in detecting biomarkers (2023, March 20)
retrieved 21 March 2023
from https://phys.org/news/2023-03-team-nanosensor-capable-single-molecule-precision.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no
part may be reproduced without the written permission. The content is provided for information purposes only.
For all the latest Science News Click Here
For the latest news and updates, follow us on Google News.

