How does an aging-associated enzyme access our genetic material?
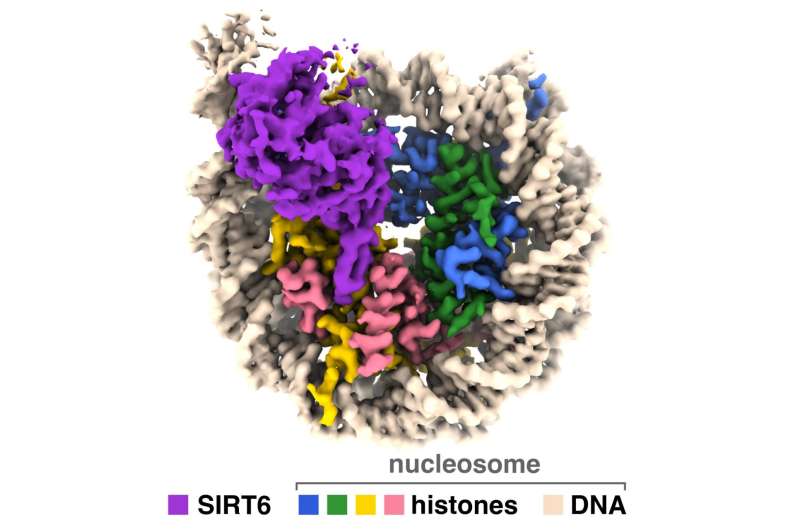
New research provides insight into how an enzyme that helps regulate aging and other metabolic processes accesses our genetic material to modulate gene expression within the cell. A team led by Penn State researchers have produced images of a sirtuin enzyme bound to a nucleosome—a tightly packed complex of DNA and proteins called histones—showing how the enzyme navigates the nucleosome complex to access both DNA and histone proteins and clarifying how it functions in humans and other animals.
A paper describing the results appears April 14 in the journal Science Advances.
Sirtuins are a type of enzyme found in organisms ranging from bacteria to humans that play important roles in aging, sensing DNA damage, and suppressing tumors in various cancers. Because of these varied roles, pharmaceutical companies are exploring their potential for biomedical applications. Much effort has focused on the ability of some sirtuins to decrease gene expression by removing a chemical flag from histone proteins.
“In our cells, DNA is not naked like we see it in textbooks; it is spooled around proteins called histones within a large complex called the nucleosome,” said Song Tan, Verne M. Willaman Professor of Molecular Biology at Penn State and an author of the paper. “This packaging can also contribute signals for turning on or turning off genes: Adding an ‘acetyl’ chemical flag to the histone packaging material turns on a gene, while removing the acetyl flag turns the gene off. Sirtuins can silence gene activity by removing the acetyl flag from histones packaged into nucleosomes. Understanding how sirtuins interact with the nucleosome to remove this flag could inform future drug discovery efforts.”
Previous studies have focused on how sirtuins interact with short segments of histones in isolation, in part because such histone “tail” peptides are much easier to work with in the lab. According to Tan, the nucleosome is a hundred times larger than typical histone peptides used in these studies and are consequently much more complicated to work with.
“We have visualized a sirtuin enzyme called SIRT6 on its physiologically relevant substrate—the entire nucleosome,” said Jean-Paul Armache, assistant professor of biochemistry and molecular biology at Penn State and an author of the paper. “And we found that SIRT6 interacts with multiple parts of the nucleosome, not only the histone where the acetyl flag is to be modified.”
Using a powerful type of imaging called cryo-electron microscopy with instruments at the Penn State Cryo-Electron Microscopy Facility, the National Cancer Institute and the Pacific Northwest Cryo-EM Center, the researchers identified how SIRT6 positions itself on the nucleosome in order to remove an acetyl group from the K9 position on the histone called H3. Following up with biochemical experiments—in collaboration with the lab of Craig Peterson at the University of Massachusetts Chan Medical School—helped confirm their results.
The researchers found that SIRT6 binds to the nucleosome using a type of connection called an “arginine anchor.” This type of binding—described by Tan’s lab in 2014—is used by a variety of proteins that target a particularly acidic patch on the nucleosome’s surface. In this case, a structural feature of SIRT6 called an extended loop nestles into a divot in the acidic patch, somewhat like a pipe sitting in a ditch.
“The arginine anchor is a common paradigm for how many chromatin proteins interact with the nucleosome,” said Tan. “When we mutated the SIRT6 arginine anchor, the activity at the K9 position was severely affected, supporting a critical role for the SIRT6’s arginine anchor. Surprisingly, this mutation also impacted SIRT6’s enzymatic activity at a different position, K56, located much further away.”
Instead of SIRT6 binding to the nucleosome in two different ways to access the two different histone positions, it is possible that SIRT6 binds to access K9 in a way that might also provide access to K56.
“SIRT6 binds to a partially unwrapped nucleosome, with DNA displaced from the end of the nucleosome” said Armache. “This exposes the K56 position, and it is possible that SIRT6 could essentially lean down to reach that position. We would like to validate this hypothesis in the future. We also hope to explore how SIRT6 works alongside other enzymes and to better understand its role in the response to DNA damage.”
More information:
Un Seng Chio et al, Cryo-EM structure of the human Sirtuin 6-nucleosome complex, Science Advances (2023). DOI: 10.1126/sciadv.adf7586
Citation:
How does an aging-associated enzyme access our genetic material? (2023, April 14)
retrieved 14 April 2023
from https://phys.org/news/2023-04-aging-associated-enzyme-access-genetic-material.html
This document is subject to copyright. Apart from any fair dealing for the purpose of private study or research, no
part may be reproduced without the written permission. The content is provided for information purposes only.
For all the latest Science News Click Here
For the latest news and updates, follow us on Google News.

